Martina Marchetti-Deschmann and Günter Allmaier
Institute of Chemical Technologies and Analytics, Vienna University of Technology, Getreidemarkt 9/164-IAC, A-1060 Vienna, Austria
Introduction
Natural latex was used by Aztec Indians to prepare shoes, bottles, balls and other products during the pre-Columbian civilisation. Latex is produced in laticifers, specialised cells located directly under the bark of the rubber tree, and is harvested as latex milk by periodic incision of the bark (tapping). Latex milk is a colloidal dispersion consisting of 30–38% rubber polymer particles, 60–70% water, 1–2% proteins, 2% resins, 1% lipids, 5–8% oligosaccharides and 0.5% inorganic salts. To prevent coagulation of freshly tapped milk, 0.7% (high ammoniated latex) or 0.2% (low ammoniated latex) ammonia is added, then the milk is centrifuged to a rubber content of >60%. Auxiliaries to inhibit fungal or bacterial infestation are admixed, and the prepared milk is shipped to different parts of the world.
More than 50% of the world production of latex milk is used for dipping products. Natural rubber latex (NRL) gloves are produced by dipping porcelain forms into a specially prepared latex milk mixture containing, inter alia, vulcanisation accelerators, anti-aging additives and lubricants. After dipping, washing (leaching) and drying procedures follow until the product is stripped off the mould. Depending on the final product, chlorination and/or starch powder treatment to prevent adherence and improve wearing comfort then follow. As more attention was turned towards wearing medical gloves to reduce the risk of infections (e.g. HIV), the occurrence of allergies related to NRL products increased significantly.1 The most common allergy type is type IV allergy, a contact dermatitis occurring 6–48 hours after contact, generally initiated by additives not completely removed. Type I allergy, a systematic allergic reaction caused by circulating IgE antibodies against proteins in NRL, was first reported in 1927.2 The first clear immediate type of allergy was reported in 1979,3 but the first evidence of a water soluble protein (MW > 30 kDa) in NRL as a potential originator for type I allergy was not reported until 1986.4
Since the early 1990s intensive investigations on latex have been undertaken, applying an array of bioanalytical methods, such as high performance liquid chromatography (HPLC), gel electrophoresis (GE), gel filtration or immunoblotting, that revealed information on 13 latex allergens (Hev b 1–13), all registered at the International Union of Immunological Societies (IUIS).5 It is commonly considered that the amount of residuals on finalised NRL products determine the allergenic potency of the products. Type I allergy-inducing chemicals are usually efficiently removed during the leaching processes and very good analytical testing methods can prove the gloves’ general compliance with regulations.6 However, the accurate measurement of protein levels in order to evaluate type IV allergy potential is still a challenge as both regulative approaches, total protein content7 and immunological tests,8 depend highly on a comprehensive protein extraction procedure. Direct analysis of inner and outer latex glove surfaces using matrix-assisted laser desorption/ionisation ion mobility quadrupole time-of-flight mass spectrometry (MALDI-IM-QToF-MS) to detect allergens present was expected to help to overcome, at least partly, the problem of incomplete protein extraction/detection. MS results have been compared to standard bioanalytical methods using HPLC-UV detection and SDS-PAGE combined with immunoblotting.
Methods
Different types of commercially available NRL gloves were cut and ground up to pieces no larger than 2 × 2 mm. 0.4 g of the powder was extracted with 1 mL sample buffer for GE according to Lämmli9 by boiling. For protein enrichment/purification ultrafiltration was applied. 25 µL of the enriched extract was analysed by reversed-phase (C18) HPLC applying a trifluoroacetic acid (TFA) in water/acetonitrile gradient. The amino acid (AA) content of each HPLC fraction was also determined.10 The total NRL glove extract and single fractions of the HPLC separation were separated by GE for fast and easy protein detection. Full experimental details can be found in References 10–12. For direct NRL glove analysis, an analysis circumventing the critical protein extraction, MALDI-IM-QToF MS (Waters, Synapt HDMS), was applied. For this the gloves were rinsed with water to remove any unwanted residues (e.g. starch) and chopped into small pieces (approximately 2 × 2 mm). These pieces were fixed on a MALDI target (see later, Figure 4A) with the side of interest up using double-sided adhesive tape. After fixation the glove surfaces were etched with 0.5 µL TFA and dried at room temperature. Then the MALDI matrix was applied. For best results a special matrix mixture had to be used.11 MALDI mass spectra (m/z range: 4000–16,000) were acquired, rastering over a surface area of 150 µm × 150 µm accumulating 1000 unselected mass spectra.
Results
As shown in Figure 1A a lot of signals could be detected after HPLC-UV analysis of the NRL glove extract demonstrating the very complex composition of a finalised latex product. Sample buffer ingredients could easily be assigned (highlighted in yellow in Figure 1A) and most of the remaining signals were further assigned to vulcanisation auxiliaries, accelerators or lubricants.12 As NRL allergens were the main focus of the study and proteins are generally easily detected after GE separation and Coomassie staining, the total glove extract and single HPLC fractions, collected every minute (assigned by the corresponding fraction number), were analysed on 16% Tricine gels (Figure 1B). In fraction 25 a faint smear between 3 kDa and 14.2 kDa was detected but evidence for the presence of NRL allergens, distinguishable by clear blue colouring of the protein band, was not found before fractions 29–31 (broad band at 5 kDa) and 34–36 (two distinct bands at 14 kDa). Quantitative AA analysis (20 commonly occurring AAs) ruled out that the faint band in fraction 25 was a protein but corroborated the presence of proteins in fractions 29–31 and 34–38 (Table 1). The protein bands also showed positive immunological reactions (moderate for fraction 29–31, strong for fraction 34–36) towards polyclonal Hev b 1 antibodies.10 T-cell proliferation tests using patient sera of persons with a history of NRL allergy confirmed the findings of allergens still present in finalised NRL glove products.10 The two distinct bands in fraction 34–36 were previously identified applying state-of-the-art proteomics tools, i.e. database search of identified sequence tags after in-gel digestion and MS experiments.13
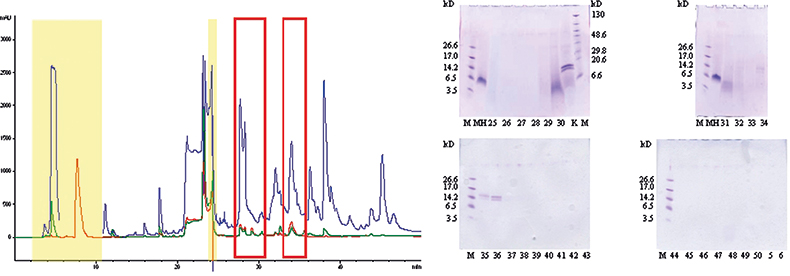
Figure 1. (A) 25 µL of total natural rubber latex glove extract (in electrophoresis buffer) separated by reversed-phase chromatography. UV—215 nm (blue), 254 nm (green) and 280 nm (red). Yellow box—buffer components, red rectangles—protein containing HPLC fractions, other detected components—auxiliaries added during production process. (B) Gel electrophoretic separation of the total NRL glove extract (K) and single HPLC fractions.
Table 1. Quantitative amino acid content by HPLC.10
HPLC fraction | Amino acid content (µg /100 µL) |
8–16 | 1.77 |
17–18 | 1.16 |
21–22 | 3.21 |
23–25 | 3.40 |
29–31 | 54.28 |
32–33 | 7.70 |
34–38 | 15.58 |
39–40 | 3.01 |
41–43 | 1.96 |
44–46 | 0.51 |
47–48 | 0.06 |
49–50 | 0.19 |
The upper gel band at 14.6 kDa represents Hev b 1, the lower a protein at 13.6 kDa which is a 10 amino acids shorter form of Hev b 1. The proteomics approach was not successful for fraction 29–31 and therefore they were of certain interest. As it is known that Hev b 6, the probably best studied NRL allergen (20 kDa), is processed into a 5 kDa N-terminal domain [Hev b 6.02, monoisotopic (m.i.) molecular mass 4723.82 Da] and a 14 kDa C-domain (Hev b 6.03) both showing high clinical reactivity, the protein determined in fraction 29–31 after GE was from this point suspected to be Hev b 6.02. The detection of all, up to this point, identified NRL allergens in addition to the detection of the 5 kDa component without any potential modification was of interest.
All analytical methods reported in the literature and described above have a high potential to alter NRL allergens present in the gloves, e.g. by hydrolysis (degradation), oxidation or side chain modification. It is then no longer clearly distinguishable if the protein modification arises from the manufacturing process or from sample preparation. Therefore the explanation for the detected protein bands at 13.6 kDa and 14.6 kDa after GE could either be sample preparation artefacts or that protein truncation has really taken place during the manufacturing process. So, direct determination of the accurate molecular weight (MW) of all present proteins in finalised NRL gloves without complex sample preparation was a main issue. In addition, the development of an analytical method for a fast and easy in situ localisation of NRL allergens in gloves, e.g. to estimate the allergenic potential of the latex material, was desirable. A sample preparation protocol was developed applying a binary MALDI matrix11 on TFA-etched latex glove surfaces allowing the direct determination of different protein components on NRL glove surfaces by means of a MALDI-IM-QToF mass spectrometer (Figure 2A). A prominent component at m/z 4718.034 (m.i.; [M + H]+) with minor accompanying analytes was detected on the inner surface of the latex gloves—the surface in contact with, for example, the healthcare worker but not the patient. Additionally the already clearly identified components at 13.6 kDa (truncated Hev b 1) and 14.6 kDa (Hev b 1) could be detected. The high mass resolution achieved with the spectrometer in this m/z range made the detection of m.i. m/z values possible, leading to the detection of a number of Hev b 6 variants and unambiguous assignments of certain AA elongations. Figure 2B shows the assignment of certain sequence variations of m/z 4718.034 [addition of Serine (S), Glycine (G) and Glutamic Acid (E)] only partially described previously.11,14 The mass difference between the two major components, m/z 4718.034 and m/z 4699.997, could be determined to be 18.037 Da (Figure 2C). The most probable explanation for this observation is a pyroglutamic acid formation (E → Pyro-E, theoretical m.i. mass difference: –18.011 Da). Pyro-E formation is a commonly occurring AA modification, either naturally or introduced during sample preparation, which is, in the case of NRL glove surfaces, the manufacturing process or MALDI sample preparation itself. All mentioned sequence modifications (+S-G-E, pyro-E) are in good agreement with the sequence published in the literature if m/z 4718.034 is accepted to be Hev b 6 (Figure 3). The first AA of the protein sequence is E, therefore pyro-E formation is very likely. The detection of signals hinting at the presence of additional AAs (S, G, E) point to the fact that the determined compound is not the biologically processed form of the protein, the N-terminus Hev b 6.02, but a degradation product of Hevein, Hev b 6 (23 kDa), present in freshly tapped NRL milk. The determined MW for the major component, 4717.034 Da, is not in total agreement with the published data for Hev b 6.02. A mass difference of –6.786 Da can be calculated. The fact that a sequence variation unlikely to be observed for the biologically processed Hev b 6.02 was detected, means that in all probability the major component originates from hydrolysed Hev b 6. This makes the assumption of further, up to now not known, sequence modifications very probable.

Figure 2. (A) Positive-ion MALDI mass spectrum recorded on a MALDI-IM-QToF-MS instrument directly from the inner surface of a NRL glove. (B) Detailed view of the m/z range related to the N-terminus of Hevein (major latex allergen). A sequence elongation could clearly be assigned. (C) The high mass resolution allows the assignment of mass differences with very high mass accuracy to detect potential pyro-glutamic acid formation (theoretical monoisotopic mass difference of 18.011 Da).

Figure 3. Amino acid sequence of Hevein, Hev b 6 and its substructures. Red rectangles—glutamic acid prone to pyroglutamic acid formation and three amino acids detected by MALDI-IM-QToF-MS elongating mature Hev b 6.02.
The production process, dipping of a ceramic form into NRL milk, followed by drying, extensive leaching processes and stripping from the mould, thereby turning the product inside out, results in the inner and outer surfaces of NRL gloves not being identical. Direct analysis of the NRL glove surfaces was corroborating this assumption. The intensity of the MS signals between m/z 4718 and m/z 4727 (m.i. distribution for the major component) was significantly higher on the inner than on the outer surface (Figure 4B). Furthermore, performing so-called MALDI imaging experiments —the assignment of detected m/z values to certain locations on the investigated NRL glove surface and by transforming this data set into an image—enables the spatial distribution of analytes to be studied in detail (Figure 4B).
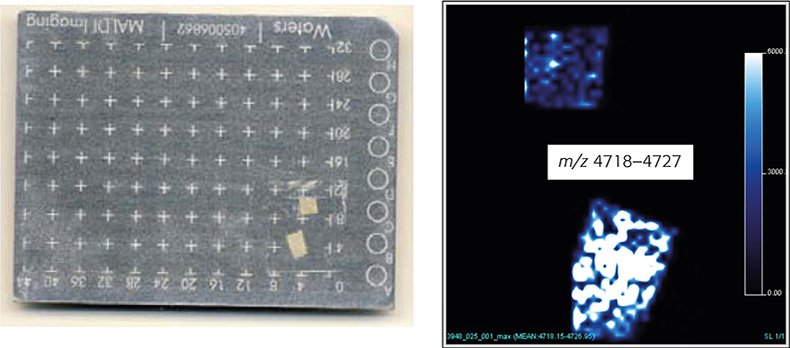
Figure 4. MALDI MS image: (A) 2 × 2 mm glove pieces attached to a MALDI sample plate. The smaller piece represents the outer surface of a NRL glove, the larger piece the inner surface. (B) Intensity plots for the distribution of the most abundant surface associated component, a degradation product of Hev b 6.
Conclusions
MALDI high resolution mass spectrometry demonstrated directly that Hev b 6 and Hev b 1, and also truncated forms thereof, are present on the inner surface of medical NRL gloves. The speed and high accuracy of the applied method and instrumentation makes the detection of surface associated proteins feasible without any prior protein extraction procedure (in situ localisation). This is a very important fact as artificial protein alteration has to be prevented. The reported example demonstrates the direct MALDI characterisation and MALDI imaging mass spectrometry of industrial material surfaces originating from a natural source after processing.
Acknowledgements
We thank Joanna Kirkpatrick and Matt Kennedy for the help with the Synapt HD mass spectrometer.
References
- B.J. Meade, D.N. Weissman and D.H. Beezhold, Int. Immunopharmacol. 2, 225–238 (2002). https://doi.org/10.1016/S1567-5769(01)00175-8
- G. Stern, Klin. Wochenschr. 6, 1096–1097 (1927). https://doi.org/10.1007/BF01890315
- A.F. Nutter, Brit. J. Dermatol. 101, 597–598 (1979). https://doi.org/10.1111/j.1365-2133.1979.tb11893.x
- T. Carillo, M. Cuevas, T. Munoz, M. Hinojosa and I. Moneo, Contact Dermatitis 15, 69–72 (1986). https://doi.org/10.1111/j.1600-0536.1986.tb01279.x
- H.Y. Yeang, Curr. Opin. Allergy and Clin. Immunol. 4, 99–104 (2004). https://doi.org/10.1097/00130832-200404000-00005
- ASTM International, “ASTM D7103–06: Standard Guide for Assessment of Medical Gloves”, in A.S.F.T.A. Materials. West Conshohocken, PA, USA (2006).
- ASTM International, “ASTM 5712–95: Analysis of Protein in Natural Rubber and Its Products”, in A.S.F.T.A. Materials. West Conshohocken, PA, USA (1995).
- ASTM International, “ASTM D6499–00: Standard Method for The Immunological Measurement of Antigenic Protein in Natural Rubber and its Products”, in A.S.F.T.A. Materials. West Conshohocken, PA, USA (2003).
- U.K. Lämmli, Nature 227, 680–685 (1970). https://doi.org/10.1038/227680a0.
- Dürauer, Characterization of allergenic proteins in natural latex gloves. Department of Biotechnology, University of Natural Resources and Applied Life Sciences, Vienna, Austria, PhD thesis, p. 204 (2001).
- M. Marchetti-Deschmann and G. Allmaier, J. Mass Spectrom. 44, 61–70 (2009). https://doi.org/10.1002/jms.1471.
- M. Marchetti, Characterization of allergy related proteins and peptides in natural latex gloves and flowers of Sambucus nigra by mass spectrometry. University of Vienna, Vienna, Austria, PhD thesis, p. 174 (2002).
- Dürauer, E. Csaszar, K. Mechtler, A. Jungbauer and E. Schmid, J. Chromatogr. A 890, 145–158 (2000). https://doi.org/10.1016/S0021-9673(00)00241-7.
- U.M. Soedjanaatmadja, J. Hofsteenge, C.M. Jeronimus-Stratingh, A.P. Bruins and J.J. Beintema, Biochim. Biophys. Acta 1209, 144–148 (1994). https://doi.org/10.1016/0167-4838(94)90150-3





